Load the Data
library(covdata)
#>
#> Attaching package: 'covdata'
#> The following object is masked from 'package:datasets':
#>
#> uspop
library(tidyverse)
#> ── Attaching packages
#> ───────────────────────────────────────
#> tidyverse 1.3.2 ──
#> ✔ ggplot2 3.4.0 ✔ purrr 1.0.1
#> ✔ tibble 3.1.8 ✔ dplyr 1.0.10
#> ✔ tidyr 1.2.1 ✔ stringr 1.5.0
#> ✔ readr 2.1.3 ✔ forcats 0.5.2
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()National level case and mortality data from the European Centers for Disease Control.
covnat_weekly
#> # A tibble: 4,966 × 11
#> date year_week cname iso3 pop cases deaths cu_ca…¹ cu_de…² r14_c…³
#> <date> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 2019-12-30 2020-01 Austr… AUT 8.93e6 NA NA NA NA NA
#> 2 2020-01-06 2020-02 Austr… AUT 8.93e6 NA NA NA NA NA
#> 3 2020-01-13 2020-03 Austr… AUT 8.93e6 NA NA NA NA NA
#> 4 2020-01-20 2020-04 Austr… AUT 8.93e6 NA NA NA NA NA
#> 5 2020-01-27 2020-05 Austr… AUT 8.93e6 NA NA NA NA NA
#> 6 2020-02-03 2020-06 Austr… AUT 8.93e6 NA NA NA NA NA
#> 7 2020-02-10 2020-07 Austr… AUT 8.93e6 NA NA NA NA NA
#> 8 2020-02-17 2020-08 Austr… AUT 8.93e6 NA NA NA NA NA
#> 9 2020-02-24 2020-09 Austr… AUT 8.93e6 12 0 12 0 NA
#> 10 2020-03-02 2020-10 Austr… AUT 8.93e6 115 0 127 0 1.42
#> # … with 4,956 more rows, 1 more variable: r14_deaths <dbl>, and abbreviated
#> # variable names ¹cu_cases, ²cu_deaths, ³r14_casesPlot national cases over time, highlighting specific countries of interest
## Libraries for the graphs
library(ggrepel)
## Convenince "Not in" operator
"%nin%" <- function(x, y) {
return( !(x %in% y) )
}
## Countries to highlight
focus_on <- covnat_weekly %>%
filter(cu_cases > 99, pop > 1e7) %>%
mutate(rate = cu_cases / pop) %>%
group_by(iso3) %>%
summarize(mean_rate = mean(rate, na.rm = TRUE)) %>%
slice_max(n = 20, order_by = mean_rate) %>%
pull(iso3)
## Colors
cgroup_cols <- c("#195F90FF", "#D76500FF", "#238023FF", "#AB1F20FF", "#7747A3FF",
"#70453CFF", "#D73EA8FF", "#666666FF", "#96971BFF", "#1298A6FF",
"#6F9BD6FF", "#FF952DFF", "#195F90FF", "#D76500FF", "#238023FF",
"#70453CFF", "#D73EA8FF", "#666666FF", "#96971BFF", "#1298A6FF",
"gray70")
covnat_weekly %>%
filter(cu_cases > 99, pop > 1e7) %>%
mutate(rate = cu_cases / pop) %>%
mutate(days_elapsed = date - min(date),
end_label = ifelse(date == max(date), cname, NA),
end_label = recode(end_label, `United States` = "USA",
`Bolivia, Plurinational State of` = "Bolivia",
`Russian Federation` = "Russia",
`Dominican Republic` = "Dominican Rep.",
`Iran, Islamic Republic of` = "Iran",
`Korea, Republic of` = "South Korea",
`United Kingdom` = "UK"),
cname = recode(cname, `United States` = "USA",
`Iran, Islamic Republic of` = "Iran",
`Korea, Republic of` = "South Korea",
`United Kingdom` = "UK"),
end_label = case_when(iso3 %in% focus_on ~ end_label,
TRUE ~ NA_character_),
cgroup = case_when(iso3 %in% focus_on ~ iso3,
TRUE ~ "ZZOTHER")) %>%
ggplot(mapping = aes(x = days_elapsed, y = cu_cases / pop,
color = cgroup, label = end_label,
group = cname)) +
geom_line(size = 0.5) +
geom_text_repel(nudge_x = 0.75,
segment.color = NA) +
guides(color = FALSE) +
scale_color_manual(values = cgroup_cols) +
labs(x = "Days Since 100th Confirmed Case",
y = "Cumulative reported Cases per capita",
title = "Cumulative Reported Cases of COVID-19 per capita",
subtitle = paste("Selected Countries with >1m population. ECDC data as of", format(max(covnat_weekly$date), "%A, %B %e, %Y")),
caption = "Kieran Healy @kjhealy / Data: https://www.ecdc.europa.eu/") +
theme_minimal()
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> Warning: The `<scale>` argument of `guides()` cannot be `FALSE`. Use "none" instead as
#> of ggplot2 3.3.4.
#> Don't know how to automatically pick scale for object of type <difftime>.
#> Defaulting to continuous.
#> Warning: Removed 1940 rows containing missing values
#> (`geom_text_repel()`).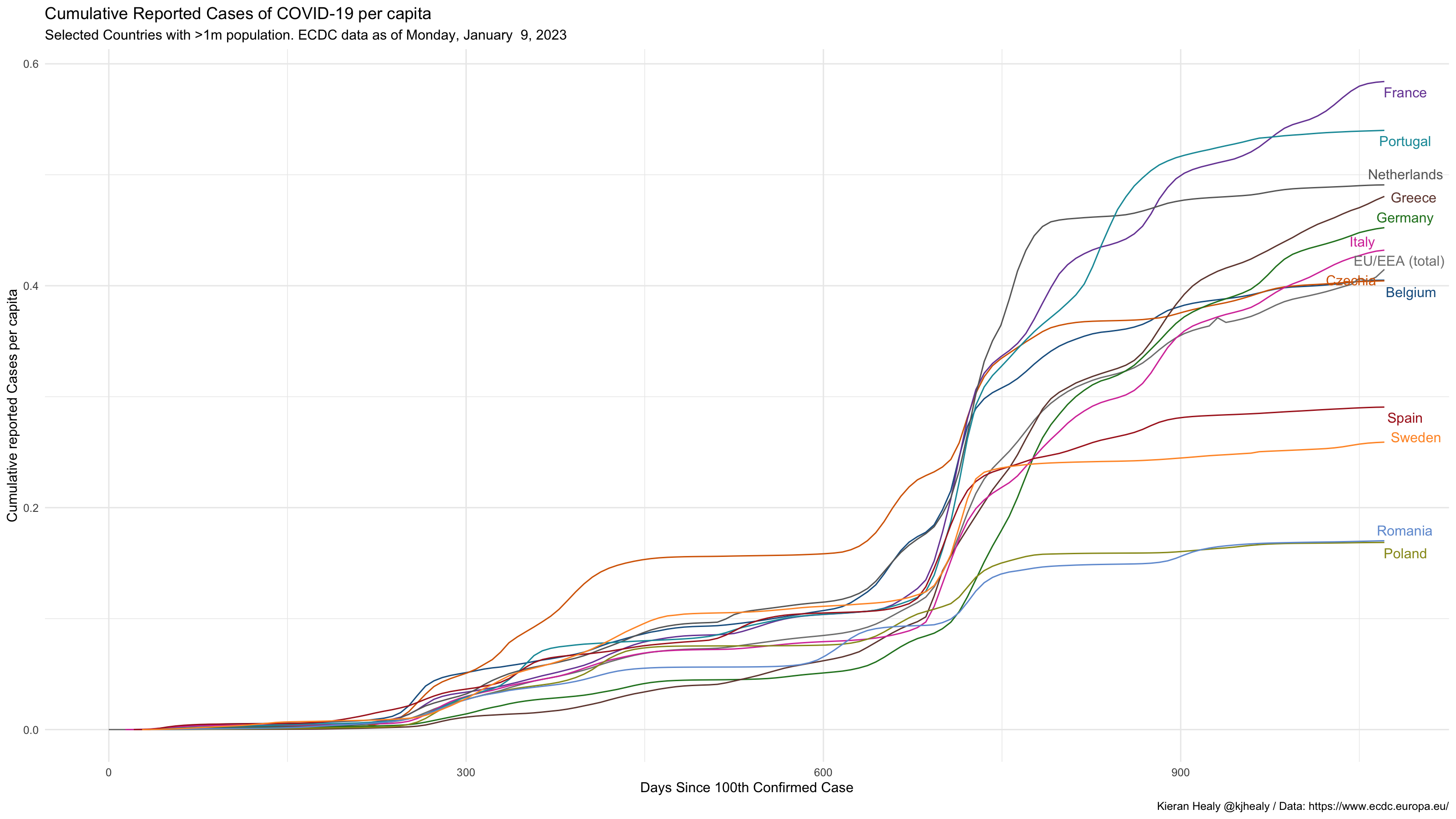
Deaths per Capita
covnat_weekly %>%
filter(iso3 %in% focus_on) %>%
ggplot(mapping = aes(x = date, y = (deaths/pop)*1e6)) +
geom_line(size = 0.5) +
# scale_y_continuous(labels = scales::comma_format(accuracy = 1)) +
facet_wrap(~ cname) +
labs(x = "Date",
y = "Deaths per Million Population",
title = "Deaths from COVID-19, Selected Countries",
subtitle = paste("ECDC weekly data as of", format(max(covnat_weekly$date), "%A, %B %e, %Y")),
caption = "Kieran Healy @kjhealy / Data: https://www.ecdc.europa.eu/") +
theme_minimal()
#> Warning: Removed 4 rows containing missing values (`geom_line()`).
Note the effects of reporting issues in various countries.